Note
Go to the end to download the full example code.
Use result helpers to load custom data#
The Result class, which is an instance
created by the Model, gives
access to helpers for requesting results on specific mesh and time scopings.
With these helpers, working on a custom spatial and temporal subset of the
model is straightforward.
# Import necessary modules
from ansys.dpf import core as dpf
from ansys.dpf.core import examples
Create a model object to establish a connection with an example result file:
model = dpf.Model(examples.download_multi_stage_cyclic_result())
print(model)
DPF Model
------------------------------
Modal analysis
Unit system: MKS: m, kg, N, s, V, A, degC
Physics Type: Mechanical
Available results:
- node_orientations: Nodal Node Euler Angles
- displacement: Nodal Displacement
- stress: ElementalNodal Stress
- elastic_strain: ElementalNodal Strain
- elastic_strain_eqv: ElementalNodal Strain eqv
- element_orientations: ElementalNodal Element Euler Angles
- structural_temperature: ElementalNodal Structural temperature
------------------------------
DPF Meshed Region:
3595 nodes
1557 elements
Unit: m
With solid (3D) elements
------------------------------
DPF Time/Freq Support:
Number of sets: 6
Cumulative Frequency (Hz) LoadStep Substep Harmonic index
1 188.385357 1 1 0.000000
2 325.126418 1 2 0.000000
3 595.320548 1 3 0.000000
4 638.189511 1 4 0.000000
5 775.669703 1 5 0.000000
6 928.278013 1 6 0.000000
Visualize specific mode shapes#
Choose the modes to visualize:
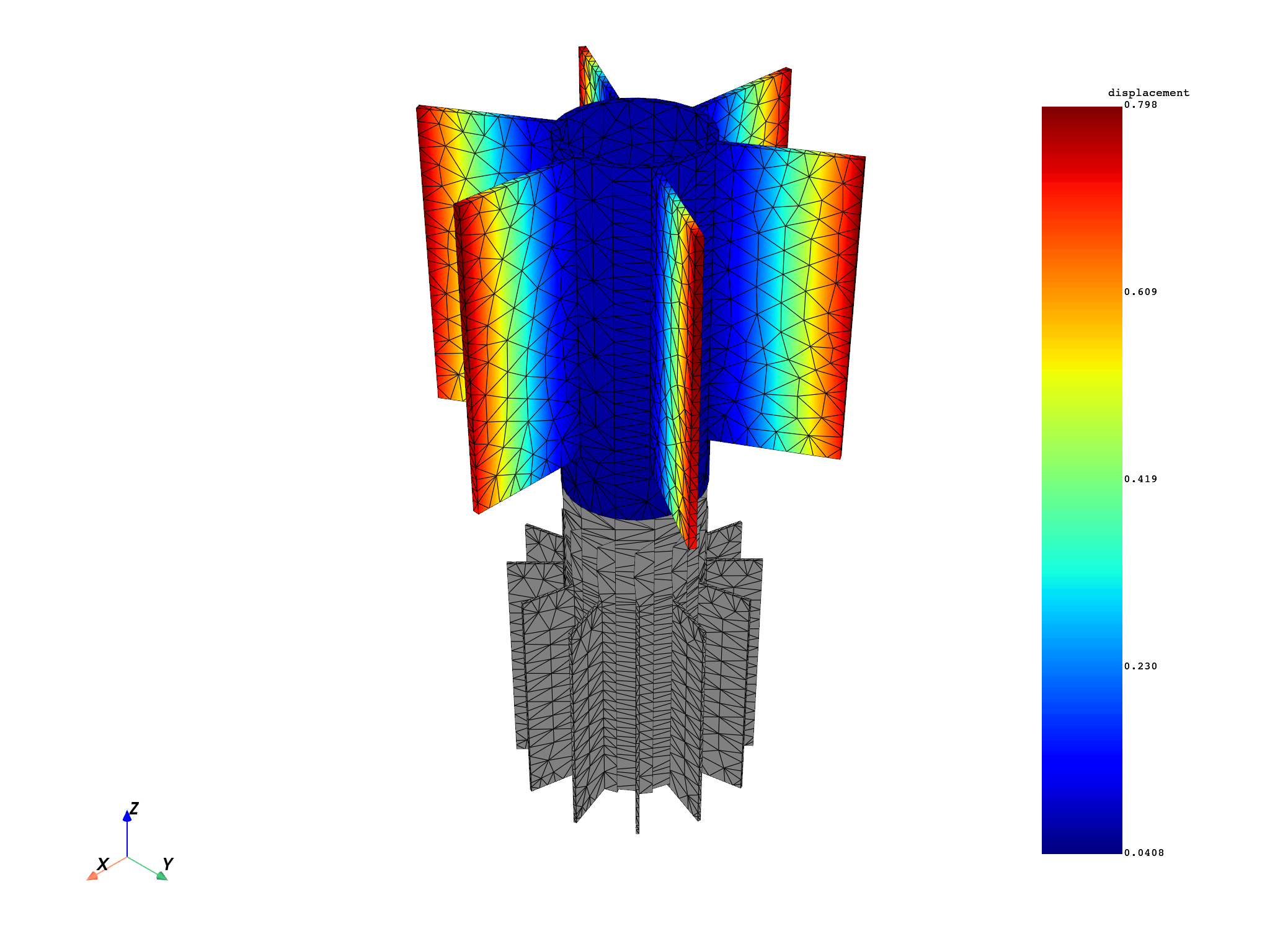
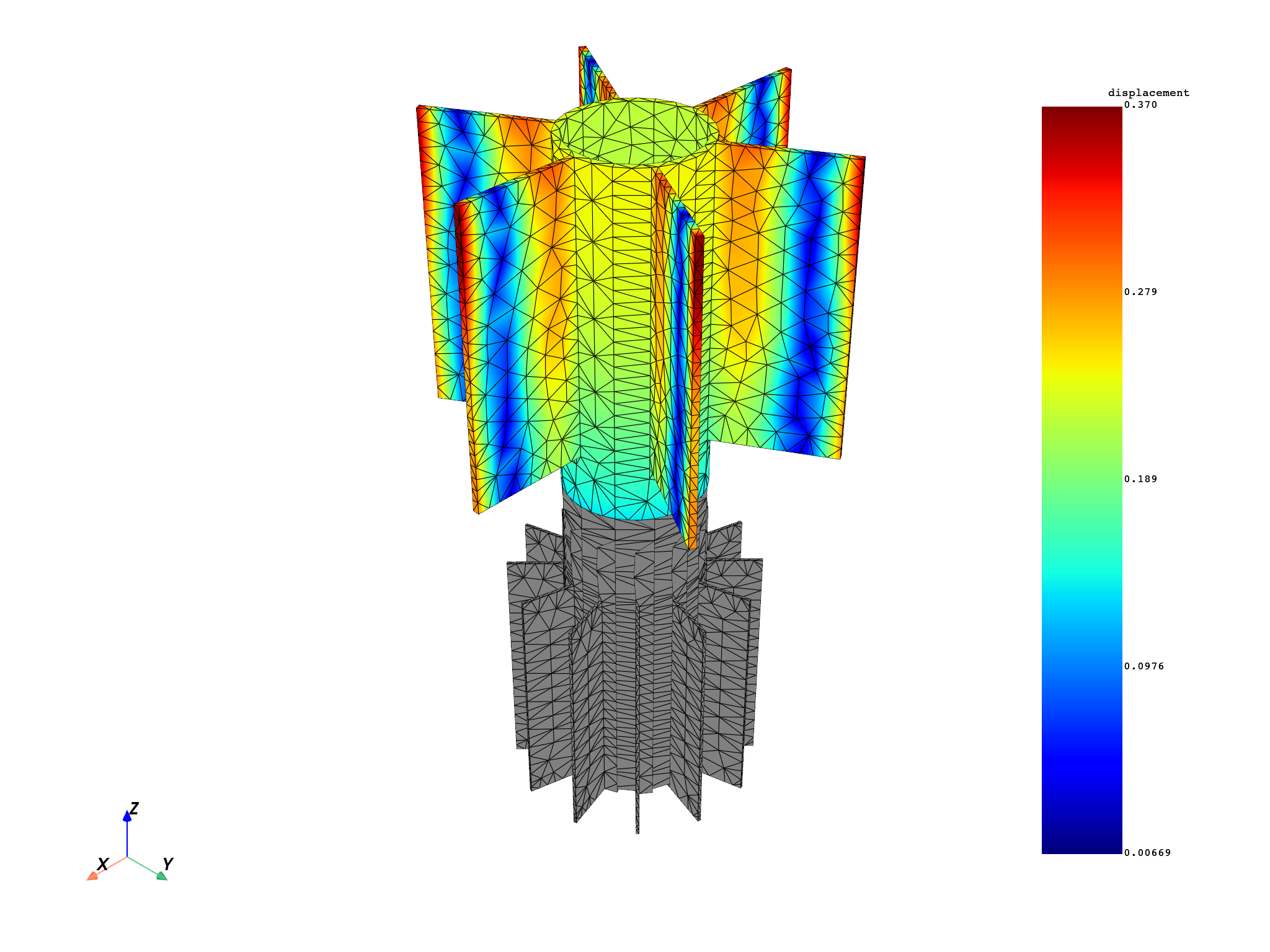
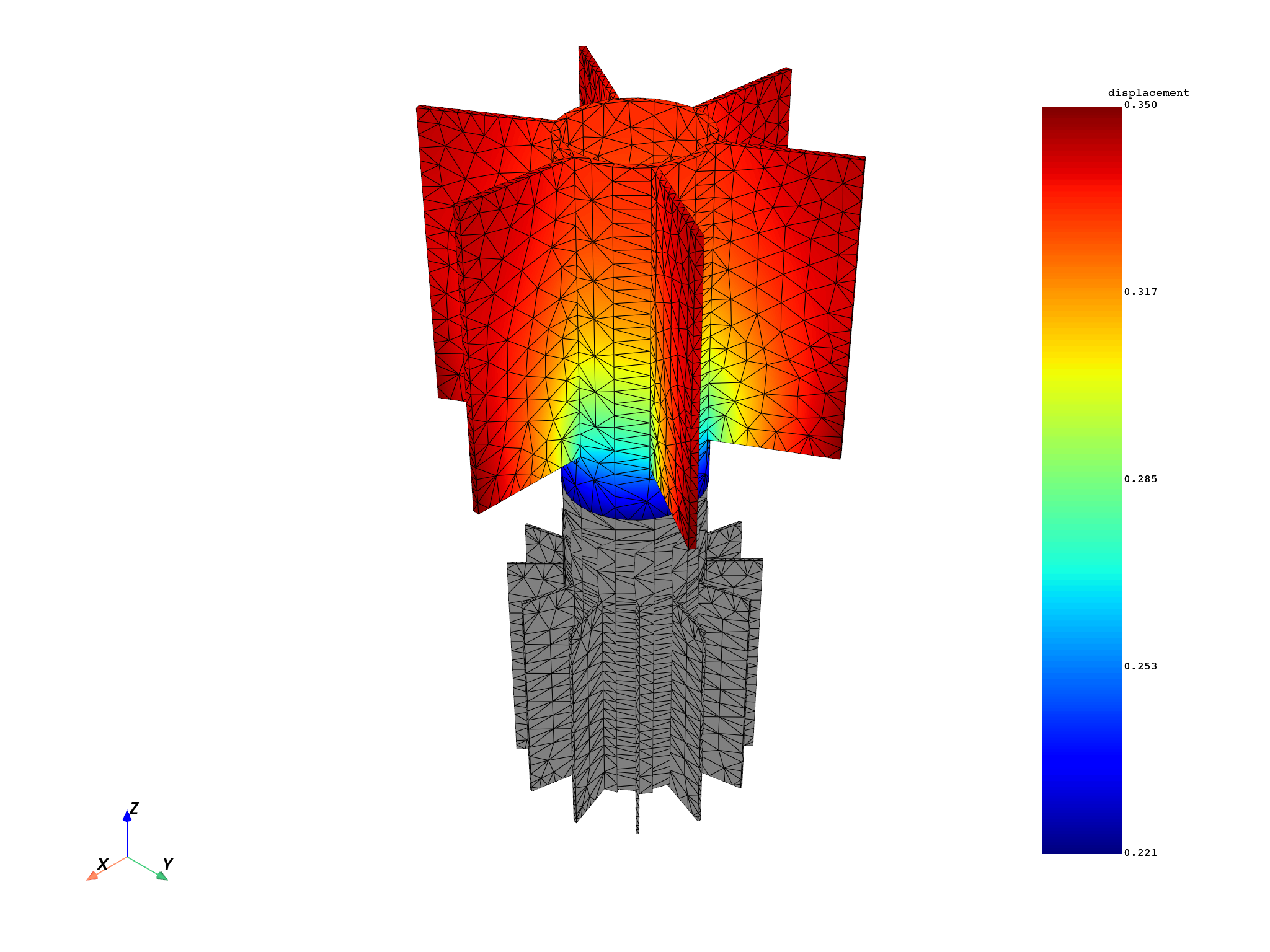
modes = [1, 5, 6]
disp = model.results.displacement.on_time_scoping(modes)
Choose a spatial subset#
Work on only a named selection (or component).
Print the available named selection:
print(model.metadata.available_named_selections)
['BC', 'ELM', 'STAG1', 'STAG1HIGH', 'STAG1LOW', 'STAG2', 'STAG2HIGH', 'STAG2LOW', '_BC_NOD', '_FIXEDSU', '_INTF_ELM', '_INTF_NOD', '_NOD', '_STAG1_BASE_ELM', '_STAG1_BASE_NOD', '_STAG1_CYCHIGH_NOD', '_STAG1_CYCLOW_NOD', '_STAG2_BASE_ELM', '_STAG2_BASE_NOD', '_STAG2_CYCHIGH_NOD', '_STAG2_CYCLOW_NOD']
Specify to the result that you want to work on a specific named selection:
disp.on_named_selection("_STAG1_BASE_NOD")
op = disp()
op.inputs.read_cyclic(2) # expand cyclic
results = op.outputs.fields_container()
# plot
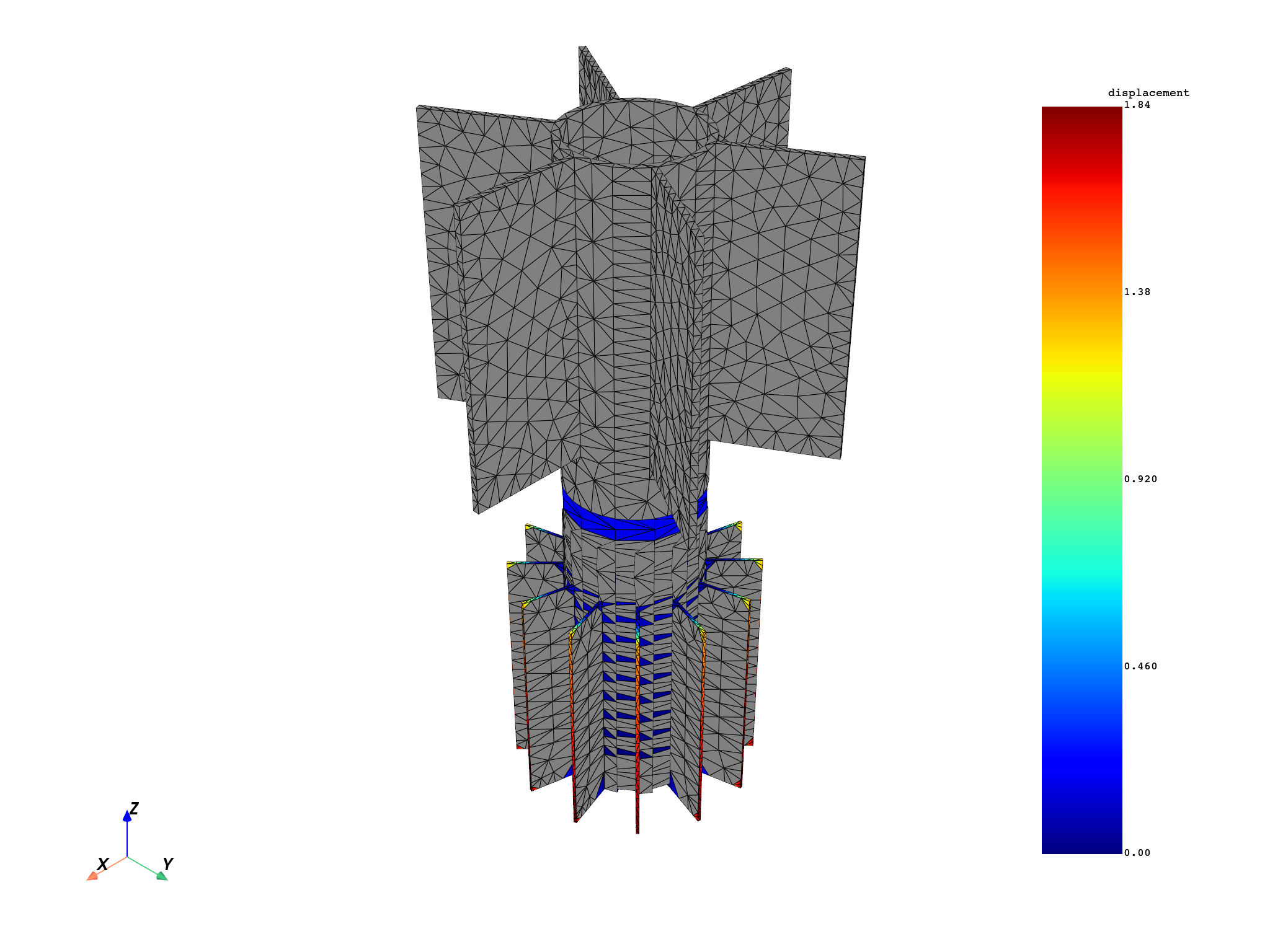
for mode in modes:
results[0].meshed_region.plot(results.get_fields_by_time_complex_ids(mode, 0)[0])
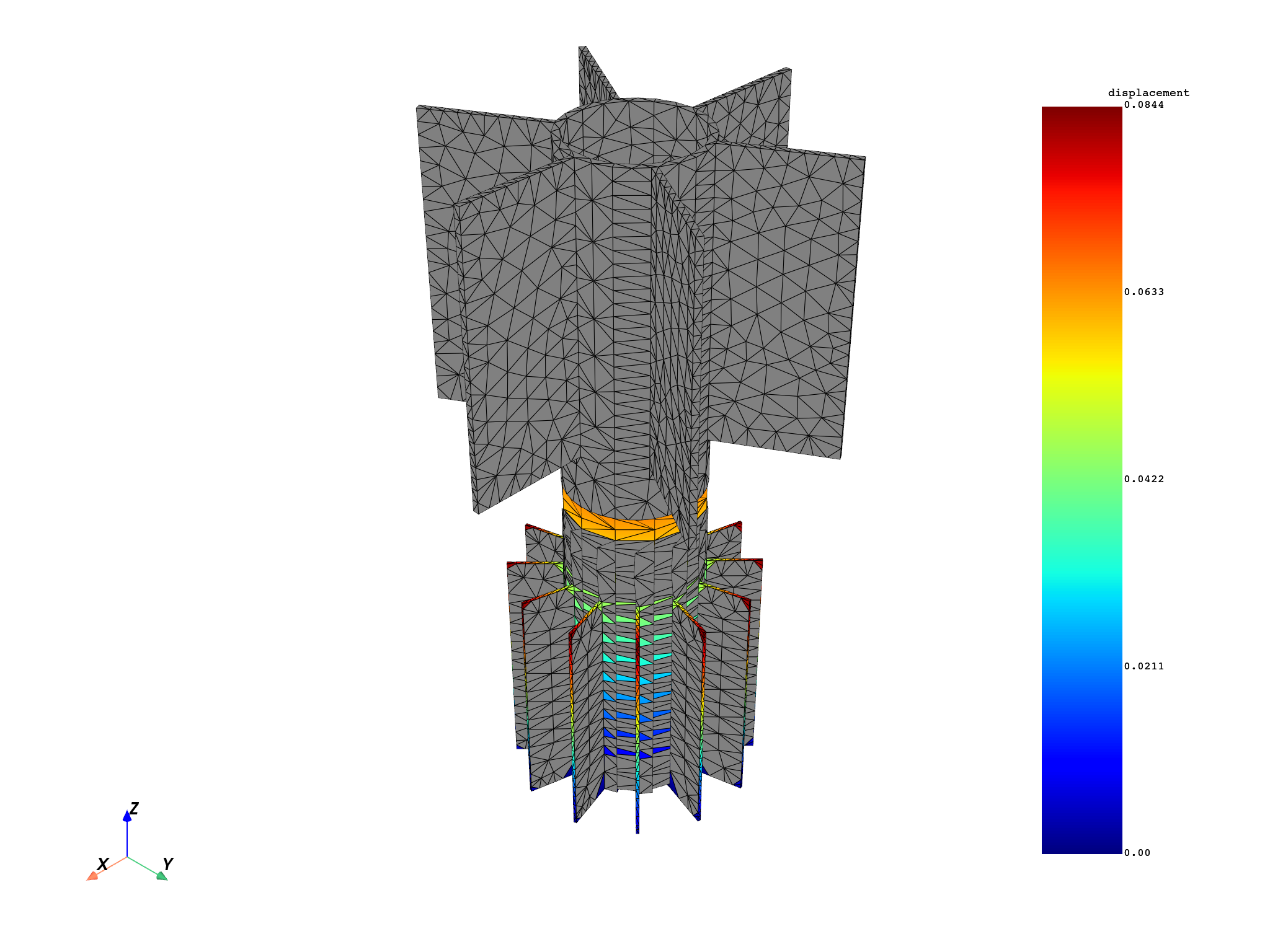
Specify to the result that you want to work on specific nodes:
disp = model.results.displacement.on_time_scoping(modes)
disp.on_mesh_scoping(list(range(1, 200)))
op = disp()
op.inputs.read_cyclic(2) # expand cyclic
results = op.outputs.fields_container()
# plot
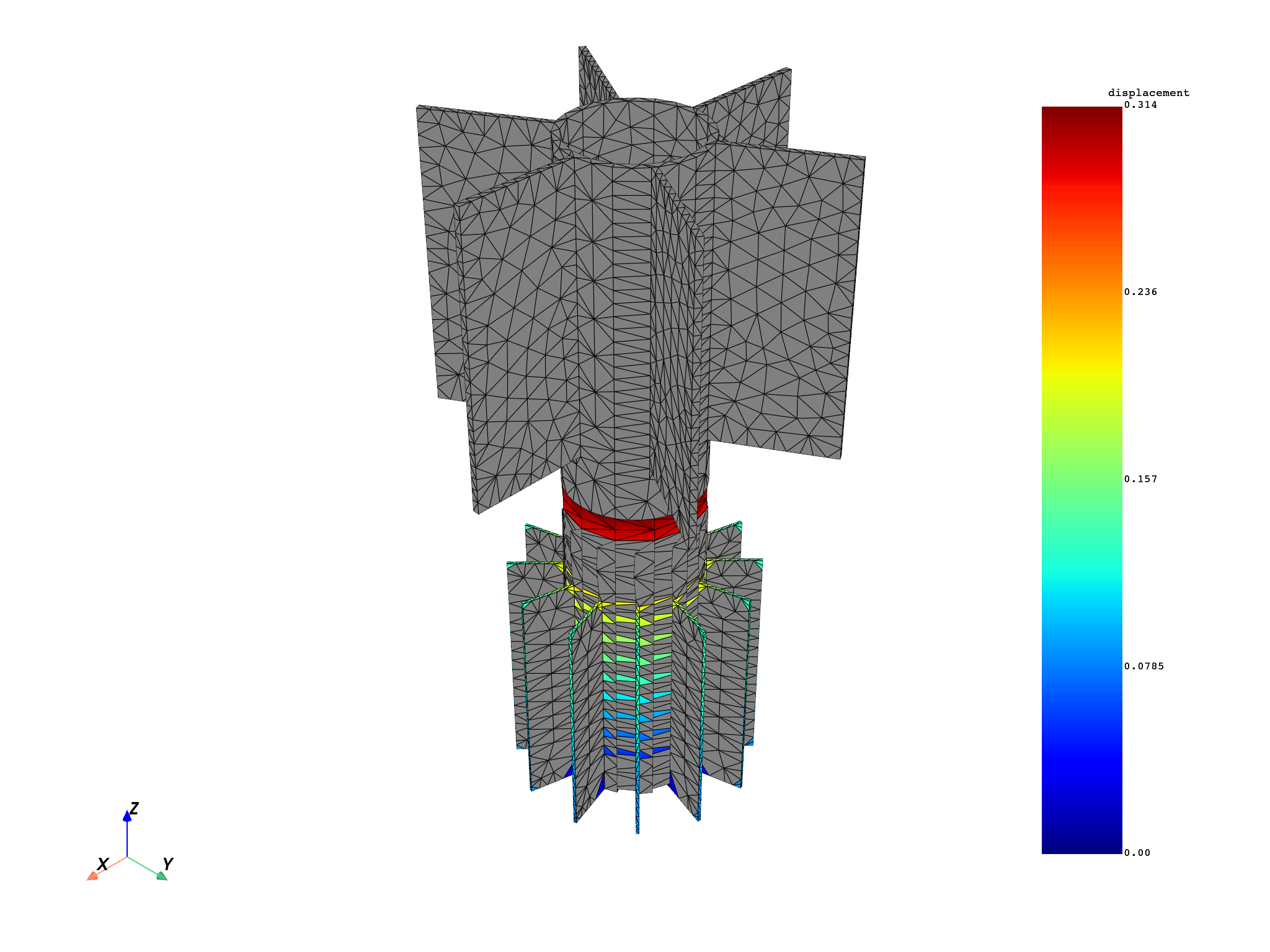
for mode in modes:
results[0].meshed_region.plot(results.get_fields_by_time_complex_ids(mode, 0)[0])
Total running time of the script: (0 minutes 8.266 seconds)